Difference between revisions of "Test"
| [unchecked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
|||
| (13 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | {{DISPLAYTITLE:Chronic myeloid leukaemia}} | |
| − | + | [[HAEM5:Table_of_Contents|Haematolymphoid Tumours (5th ed.)]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | {{Under Construction}} | |
| − | |||
| − | |||
| − | |||
| − | == | + | <blockquote class='blockedit'>{{Box-round|title=HAEM5 Conversion Notes|This page was converted to the new template on 2023-12-07. The original page can be found at [[HAEM4:Chronic Myeloid Leukemia (CML), BCR-ABL1 Positive]]. |
| − | + | }}</blockquote> | |
| − | |||
| − | |||
| − | |||
| − | = | + | <span style="color:#0070C0">(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use [https://www.genenames.org/ <u>HUGO-approved gene names and symbols</u>] (italicized when appropriate), [https://varnomen.hgvs.org/ HGVS-based nomenclature for variants], as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click nearby within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see </span><u>[[Author_Instructions]]</u><span style="color:#0070C0"> and [[Frequently Asked Questions (FAQs)|<u>FAQs</u>]] as well as contact your [[Leadership|<u>Associate Editor</u>]] or [mailto:CCGA@cancergenomics.org <u>Technical Support</u>])</span> |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | ==Primary Author(s)*== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Jack Reid, MD (University of California, Irvine) | |
| − | + | Mark Evans, MD (University of California, Irvine) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Fabiola Quintero-Rivera, MD (University of California, Irvine) | |
| − | |||
| − | + | __TOC__ | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Cancer Category / Type== | |
| − | + | Myeloproliferative neoplasm | |
| − | + | ==Cancer Sub-Classification / Subtype== | |
| − | + | Not applicable. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Definition / Description of Disease== | |
| − | + | Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm that is characterized by clonal expansion of predominantly granulocytic proliferation (neutrophils, eosinophils and basophils). The majority of the patients with CML are known to have a gene rearrangement called the Philadelphia Chromosome, which is a balanced genetic translocation t(9;22)(q34.1;q11.2) involving a fusion of the Abelson gene (ABL1) from chromosome 9q34 with the breakpoint cluster region (BCR) gene on chromosome 22q11.2. Althought 80% of the clonal evolution in CML cases can be attributed to classic Ph chromosome, secondary cytogenetic aberrations can be seen such as isochromosome 17q, gain of chromosome 8 or 19. ML was first recognized in 1845<ref>{{Cite journal|last=Jm|first=Goldman|last2=Jv|first2=Melo|date=2003|title=Chronic Myeloid Leukemia--Advances in Biology and New Approaches to Treatment|url=https://pubmed.ncbi.nlm.nih.gov/14534339/|language=en|pmid=14534339}}</ref>. Nowell and Hungerford in 1960, who coined the term Philadelphia Chromosome after realizing consistent chromosomal abnormality in leukemic cells.<ref>{{Cite journal|last=Nowell|first=Peter C.|date=2007|title=Discovery of the Philadelphia chromosome: a personal perspective|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1934591/|journal=Journal of Clinical Investigation|volume=117|issue=8|pages=2033–2035|doi=10.1172/JCI31771|issn=0021-9738|pmc=1934591|pmid=17671636}}</ref> Later in 1973, the characteristic cytogenetic feature of CML was identified: reciprocal translocation of t(9;22)(q34.1;q11.2).<ref>{{Cite journal|last=Jd|first=Rowley|date=1973|title=Letter: A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia Identified by Quinacrine Fluorescence and Giemsa Staining|url=https://pubmed.ncbi.nlm.nih.gov/4126434/|language=en|pmid=4126434}}</ref> CML has the capacity to expand in both myeloid and lymphoid lineages. However, expansion is predominantly in the granulocyte compartment of the myeloid lineages in the bone marrow.<ref>{{Cite journal|last=S|first=Faderl|last2=M|first2=Talpaz|last3=Z|first3=Estrov|last4=S|first4=O'Brien|last5=R|first5=Kurzrock|last6=Hm|first6=Kantarjian|date=1999|title=The Biology of Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/10403855/|language=en|pmid=10403855}}</ref> In the previous WHO 4th edition, CML was be divided into 3 phases of disease: chronic phase, accelerated phase and blastic phase. Currently, in the revised classification of CML, AP at diagnosis or during treatment has been omitted and replaced by recognising only the chronic and blast phases | |
| − | + | ==Synonyms / Terminology== | |
| − | |||
| − | + | Formerly chronic myelogenous leukemia or chronic granulocytic leukemia | |
| − | |||
| − | == | + | ==Epidemiology / Prevalence== |
| − | |||
| − | + | Global annual incidence of CML is 1-2 cases per 100,000 population. CML has male predominance with male to female ratio of 1.4:1. The prevalence of CML is increasing due to successful Tyrosine kinase inhibitor (TKI) therapy. Predilection of CML among certain ethnic groups has not been reported.<ref>{{Cite journal|last=Jv|first=Melo|last2=De|first2=Gordon|last3=Nc|first3=Cross|last4=Jm|first4=Goldman|date=1993|title=The ABL-BCR Fusion Gene Is Expressed in Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/8417787/|language=en|pmid=8417787}}</ref> Regional variations in age at diagnosis and overall survival among patients with chronic myeloid leukemia from low and middle income countries.<ref>{{Cite journal|last=Am|first=Mendizabal|last2=P|first2=Garcia-Gonzalez|last3=Ph|first3=Levine|date=2013|title=Regional Variations in Age at Diagnosis and Overall Survival Among Patients With Chronic Myeloid Leukemia From Low and Middle Income Countries|url=https://pubmed.ncbi.nlm.nih.gov/23411044/|language=en|pmid=23411044}}</ref> The median age of patients diagnosed with CML is 66 years according to Surveillance, Epidemiology, and End Results (SEER) program and Medical Research Council (MRC) data. Although the etiology of CML is largely unknown, cases of CML have been reported in association with radiation exposure. No studies have shown any genetic inheritance of CML. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | == | + | ==Clinical Features== |
| − | |||
| − | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instruction: Can include references in the table. Do not delete table.'') </span> | |
| − | + | {| class="wikitable" | |
| − | + | |'''Signs and Symptoms''' | |
| − | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) | |
| − | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) | |
| + | |- | ||
| + | |'''Laboratory Findings''' | ||
| + | |<span class="blue-text">EXAMPLE:</span> Cytopenias | ||
| + | <span class="blue-text">EXAMPLE:</span> | ||
| + | {| class="wikitable" | ||
| + | |'''Signs and Symptoms''' | ||
| + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) | ||
| − | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) | |
| − | |||
| − | = | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | |||
| − | |||
| − | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) | |
| + | |- | ||
| + | |'''Laboratory Findings''' | ||
| + | |<span class="blue-text">EXAMPLE:</span> Cytopenias | ||
| − | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) | |
| + | |} | ||
| − | |||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Clinical Features|The content below was from the old template. Please incorporate above.}} | |
| − | + | Approximately 50% of the patients who are diagnosed with CML are asymptomatic and diagnosed during the routine blood tests.<ref name=":0">Silver RT. Molecular Biology of CML. In: Kufe DW, Pollock RE, Weichselbaum RR, et al., editors. Holland-Frei Cancer Medicine. 6th edition. Hamilton (ON): BC Decker; 2003. Available from: <nowiki>https://www.ncbi.nlm.nih.gov/books/NBK13554/</nowiki></ref> CML is a hematological disease that occurs predominantly in adults but in rare cases, it can occur in the pediatric population.<ref>{{Cite journal|last=Am|first=Mendizabal|last2=P|first2=Garcia-Gonzalez|last3=Ph|first3=Levine|date=2013|title=Regional Variations in Age at Diagnosis and Overall Survival Among Patients With Chronic Myeloid Leukemia From Low and Middle Income Countries|url=https://pubmed.ncbi.nlm.nih.gov/23411044/|language=en|pmid=23411044}}</ref> The onset of CML is insidious. Patients with CML usually experience dragging sensation of the abdomen due to splenomegaly. The clinical hallmark of CML is the uncontrolled proliferation of mature and maturing granulocytes at all stages of maturation: metamyelocytes, myelocytes, promyelocytes, and myeloblasts. Patients with CML usually begin with the initial chronic phase before entering the terminal blastic phase and 60-80% of the patients go through accelerated phase before reaching the terminal blastic phase. In chronic phase, CML patients show abnormal routine blood tests with clinical symptoms such as unintentional weight loss, loss of appetite, satiety, fatigue, insomnia and palpable splenomegaly. In rare cases, hyperviscosity syndrome can be a manifestation with a wide spectrum of features such as priapism, tinnitus, hearing loss, cerebral accidents and blindness. In the blastic phase, CML leukemic cells resemble acute leukemic cells morphologically. CNS and lymph node involvement are notable in the blastic phase of CML. If untreated, CML patients will progress to the terminal blastic phase in 3 to 5 years. | |
| − | |||
| − | + | </blockquote> | |
| − | + | ==Sites of Involvement== | |
| − | + | Spleen is known to be the most common site of involvement as patients with CML usually present with splenomegaly. Literature has shown that bone marrow is always involved in patients with CML. | |
| − | + | ==Morphologic Features== | |
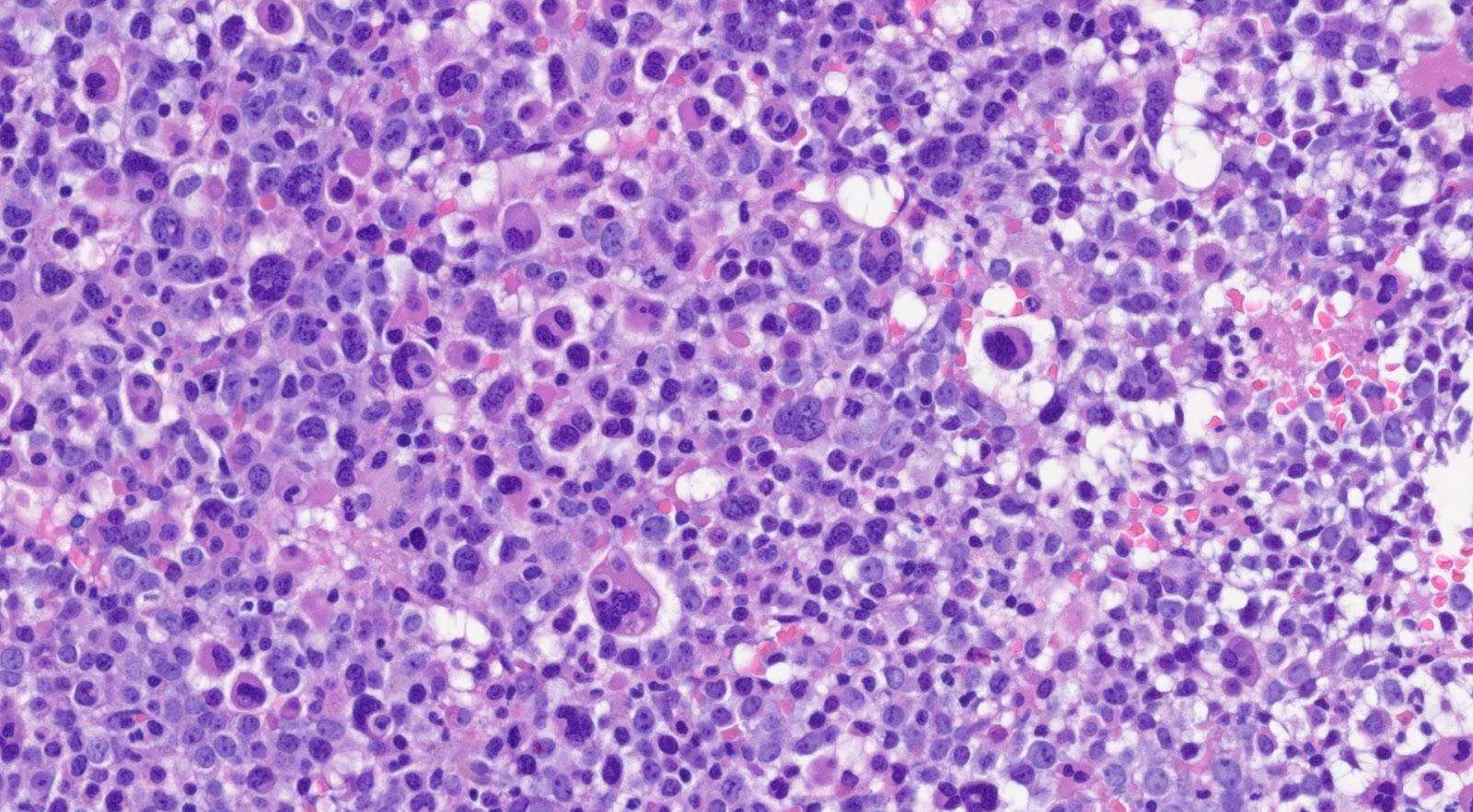
| − | + | [[File:CML 1.jpg|thumb|Chronic Myeloid Leukemia. Image courtesy of Jack Reid, MD]] | |
| − | + | Morphologically, peripheral blood smear shows the classic features of chronic-phase CML: granulocytic leukocytosis with left shift, neutrophilia, no increase in blasts, myelocyte “bulge” and basophilia.<ref>Vardiman JW, et al., (2016). Chronic myeloid leukaemia, BCR-ABL1-positive, in World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised 4th edition. Swerdlow SH, Campo E, Harris NL, et al., Editors. IARC Press: Lyon, France, p30-36.</ref> Granulocytes seen in CML patients usually lack dysplastic features. In bone marrow, the aspirate smear shows classic features of CML: marked increase in neutrophils and precursors with a myeloid: erythroid ratio of > 10:1 and small hypolobated megakaryocytes. CML can sometimes present with thrombocytosis in peripheral blood smear, mimicking essential thrombocythemia (ET). Most of the time, primary myelofibrosis (PMF) share overlapping morphological features with CML in the peripheral blood; one distinguishing feature is that the megakaryocyte morphology in PMF is large, bizarre, and hyperchromatic, which is the feature not seen in the CML. | |
| − | |||
| − | + | ==Immunophenotype== | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | The role of immunohistochemistry is minimal in diagnosing CML. CML usually shows blastic markers (CD 34, CD 117, TdT), which can be useful to confirm extramedullary (splenic involvement with blastic transformation). Lineage-specific markers (MPO, lysosome, CD42b, CD79a, PAX5, CD3) are helpful in distinguishing among myeloid, lymphoid or megakaryocytic transformation. | |
| − | + | {| class="wikitable sortable" | |
| − | + | |- | |
| + | !Finding!!Marker | ||
| + | |- | ||
| + | |Positive||CD34 | ||
| + | |- | ||
| + | |Positive||CD117 | ||
| + | |- | ||
| + | | | ||
| + | Positive | ||
| + | |Tdt | ||
| + | |} | ||
| − | + | ==Chromosomal Rearrangements (Gene Fusions)== | |
| − | |||
| − | |||
| − | |||
| − | + | Put your text here and fill in the table | |
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Chromosomal Rearrangement!!Genes in Fusion (5’ or 3’ Segments)!!Pathogenic Derivative!!Prevalence | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | ||
| + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) | ||
| + | |Yes | ||
| + | |No | ||
| + | |Yes | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | |
| + | |} | ||
| + | |||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Chromosomal Rearrangements (Gene Fusions)|The content below was from the old template. Please incorporate above.}} | |
| − | |||
| − | |||
| − | + | CML is the first cancer that is known to be linked to a specific genetic abnormality, namely the balanced chromosomal translocation known as Philadelphia Chromosome. A focal gene area of BCR (Breakpoint Cluster Region) from chromosome 22 is fused with another gene ABL (Tyrosine protein kinase ABL) that is located on chromosome 9. The chimeric oncogene BCR-ABL is the central to the pathology of CML because ABL carries a domain that is capable of phosphorylating tyrosine residues, activating a cascade of proteins that control the cell cycle. It was reported that 90% - 95% of the CML in chronic phase shows characteristic t(9;22)(q34;q11.2) reciprocal translocation that results in the Ph chromosome. This balanced translocation leads to the formation of the ''BCR/ABL'' fusion gene on chromosome 22 and a reciprocal ''ABL/BCR'' fusion gene on chromosome 9. Studies has shown that the latter gene ''ABL/BCR'' fusion gene does not seem to have any crucial role in CML and no ABL/BCR protein has been found. | |
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Chromosomal Rearrangement!!Genes in Fusion (5’ or 3’ Segments)!!Prevalence | ||
| + | |- | ||
| + | |t(9;22)(q34.1;q11.2)||3'ABL1 / 5'BCR||More than 90% | ||
| + | |} | ||
| + | |||
| + | </blockquote> | ||
| − | |||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Clinical Significance (Diagnosis, Prognosis and Therapeutic Implications).|Please incorporate this section into the relevant tables found in: | |
| + | * Chromosomal Rearrangements (Gene Fusions) | ||
| + | * Individual Region Genomic Gain/Loss/LOH | ||
| + | * Characteristic Chromosomal Patterns | ||
| + | * Gene Mutations (SNV/INDEL)}} | ||
| − | + | '''Diagnosis:''' | |
| − | + | Currently four FDA approved tyrosine kinase inhibitors (TKIs) - imatinib, nilotinib, dasatinib and bosutinib - are the first line of treatment for patients with newly diagnosed CML in chronic phase (CML-CP).<ref>{{Cite journal|last=Ja|first=Kennedy|last2=G|first2=Hobbs|date=2018|title=Tyrosine Kinase Inhibitors in the Treatment of Chronic-Phase CML: Strategies for Frontline Decision-making|url=https://pubmed.ncbi.nlm.nih.gov/29687320/|language=en|doi=10.1007/s11899-018-0449-7|pmc=PMC6023770|pmid=29687320}}</ref> For many years, inhibitors of the specific BCR-ABL1 tyrosine kinase are considered to be the most effective targeted therapy. A subset of CML patients can demonstrate resistance to TKI therapy through mutations in ABL1 and other mechanisms. The culprit of the resistance to TKI therapy can be attributed to so-called leukemic stem cells (LSCs), pluripotent BCR-ABL1+ progenitors that are largely quiescent.<ref>{{Cite journal|last=S|first=Tabarestani|last2=A|first2=Movafagh|date=2016|title=New Developments in Chronic Myeloid Leukemia: Implications for Therapy|url=https://pubmed.ncbi.nlm.nih.gov/27366312/|language=en|doi=10.17795/ijcp-3961|pmc=PMC4922205|pmid=27366312}}</ref> Therefore, understanding of signaling pathways related to survival of LSCs may be helpful. | |
| − | + | '''Prognosis''': Acquired resistance to imatinib therapy , mostly with mutation in BCR-ABL kinase domain, is known to be associated with poor prognosis.<ref>{{Cite journal|last=S|first=Branford|last2=Z|first2=Rudzki|last3=S|first3=Walsh|last4=I|first4=Parkinson|last5=A|first5=Grigg|last6=J|first6=Szer|last7=K|first7=Taylor|last8=R|first8=Herrmann|last9=Jf|first9=Seymour|date=2003|title=Detection of BCR-ABL Mutations in Patients With CML Treated With Imatinib Is Virtually Always Accompanied by Clinical Resistance, and Mutations in the ATP Phosphate-Binding Loop (P-loop) Are Associated With a Poor Prognosis|url=https://pubmed.ncbi.nlm.nih.gov/12623848/|language=en|pmid=12623848}}</ref> Five prognostic factors were shown to be associated with major cytogenetic response: the absence of blasts in peripheral blood, a hemoglobin level of more than 12 g per deciliter, the presence of less than 5 percent blasts in marrow, a time from diagnosis of CML to start of treatment of less than one year, and a history of cytogenetic relapse during interferon therapy.<ref>{{Cite journal|last=H|first=Kantarjian|last2=C|first2=Sawyers|last3=A|first3=Hochhaus|last4=F|first4=Guilhot|last5=C|first5=Schiffer|last6=C|first6=Gambacorti-Passerini|last7=D|first7=Niederwieser|last8=D|first8=Resta|last9=R|first9=Capdeville|date=2002|title=Hematologic and Cytogenetic Responses to Imatinib Mesylate in Chronic Myelogenous Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/11870241/|language=en|pmid=11870241}}</ref> | |
| − | |||
| − | + | '''Therapeutic implication''': Studies have shown that median survival | |
| − | + | Complete cytogenetic response is defined as 0% of Philadelphia-chromosome (Ph)-positive cells in metaphase in bone marrow.<ref>{{Cite journal|last=J|first=Cortes|last2=A|first2=Quintás-Cardama|last3=Hm|first3=Kantarjian|date=2011|title=Monitoring Molecular Response in Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/20960522/|language=en|doi=10.1002/cncr.25527|pmc=PMC4969001|pmid=20960522}}</ref> | |
| − | + | </blockquote> | |
| + | ==Individual Region Genomic Gain / Loss / LOH== | ||
| − | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.'') </span> | |
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Chr #!!Gain / Loss / Amp / LOH!!Minimal Region Genomic Coordinates [Genome Build]!!Minimal Region Cytoband | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | 7 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Loss | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | chr7:1-159,335,973 [hg38] | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | chr7 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Yes | ||
| + | |<span class="blue-text">EXAMPLE:</span> Yes | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | 8 | ||
| + | |<span class="blue-text">EXAMPLE:</span> Gain | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | chr8:1-145,138,636 [hg38] | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | chr8 | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Chr #!!Gain / Loss / Amp / LOH!!Minimal Region Genomic Coordinates [Genome Build]!!Minimal Region Cytoband | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | 7 | |
| + | |<span class="blue-text">EXAMPLE:</span> Loss | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | chr7:1- 159,335,973 [hg38] | |
| − | + | |<span class="blue-text">EXAMPLE:</span> | |
| − | |||
| − | |||
| − | + | chr7 | |
| + | |Yes | ||
| + | |Yes | ||
| + | |No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | |
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | 8 | |
| − | + | |<span class="blue-text">EXAMPLE:</span> Gain | |
| − | + | |<span class="blue-text">EXAMPLE:</span> | |
| − | + | chr8:1-145,138,636 [hg38] | |
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | = | + | chr8 |
| − | + | |No | |
| + | |No | ||
| + | |No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Common recurrent secondary finding for t(8;21) (add reference). | |
| + | |} | ||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Genomic Gain/Loss/LOH|The content below was from the old template. Please incorporate above.}} | |
| − | + | Not Applicable. | |
| + | </blockquote> | ||
| + | ==Characteristic Chromosomal Patterns== | ||
| − | + | Put your text here <span style="color:#0070C0">(''EXAMPLE PATTERNS: hyperdiploid; gain of odd number chromosomes including typically chromosome 1, 3, 5, 7, 11, and 17; co-deletion of 1p and 19q; complex karyotypes without characteristic genetic findings; chromothripsis. Do not delete table.'')</span> | |
| − | + | {| class="wikitable sortable" | |
| − | + | |- | |
| − | + | !Chromosomal Pattern | |
| − | + | !Diagnostic Significance (Yes, No or Unknown) | |
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | Co-deletion of 1p and 18q | ||
| + | |<span class="blue-text">EXAMPLE:</span> Yes | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Chromosomal Pattern | ||
| + | !Diagnostic Significance (Yes, No or Unknown) | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | Co-deletion of 1p and 18q | |
| + | |Yes | ||
| + | |No | ||
| + | |No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | |
| − | + | |} | |
| − | |||
| − | |||
| − | |||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Characteristic Chromosomal Aberrations / Patterns|The content below was from the old template. Please incorporate above.}} | |
| − | + | Atypical chronic myeloid leukemia (aCML) is a subtype of myelodysplastic/myeloproliferative neoplasm that lacks Philadelphia chromosome or rearrangements of PDGFRA, PDGFRB, or FGFR1. This hematological disorder has a considerable overlapping clinicopathological features with CML and CMML. It differs from CML by older median age, lower level of granulocytosis, multilineage dysplasia and lack of basophilia. Up until now, no cytogenetic changes have been associated with aCML. In peripheral blood smear, aCML typically shows granulocytic leukocytosis with striking neutrophil dysplasia (nuclear hyposegmentation and hypogranularity). | |
| − | + | </blockquote> | |
| + | ==Gene Mutations (SNV / INDEL)== | ||
| − | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: This table is not meant to be an exhaustive list; please include only genes/alterations that are recurrent and common as well either disease defining and/or clinically significant. Can include references in the table. For clinical significance, denote associations with FDA-approved therapy (not an extensive list of applicable drugs) and NCCN or other national guidelines if applicable; Can also refer to CGC workgroup tables as linked on the homepage if applicable as well as any high impact papers or reviews of gene mutations in this entity. Do not delete table.'') </span> | |
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Gene; Genetic Alteration!!'''Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other)'''!!'''Prevalence (COSMIC / TCGA / Other)'''!!'''Concomitant Mutations'''!!'''Mutually Exclusive Mutations''' | ||
| + | !'''Diagnostic Significance (Yes, No or Unknown)''' | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''TP53''; Variable LOF mutations | ||
| + | <span class="blue-text">EXAMPLE:</span> | ||
| − | + | ''EGFR''; Exon 20 mutations | |
| − | |||
| − | |||
| − | + | <span class="blue-text">EXAMPLE:</span> ''BRAF''; Activating mutations | |
| − | + | |<span class="blue-text">EXAMPLE:</span> TSG | |
| − | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | |
| − | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) | |
| − | + | |<span class="blue-text">EXAMPLE:</span> ''IDH1'' R123H | |
| + | |<span class="blue-text">EXAMPLE:</span> ''EGFR'' amplification | ||
| + | |<span class="blue-text">EXAMPLE:</span> Yes | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> No | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Gene; Genetic Alteration!!'''Presumed Mechanism (Tumor Suppressor Gene [TSG] / Oncogene / Other)'''!!'''Prevalence (COSMIC / TCGA / Other)'''!!'''Concomitant Mutations'''!!'''Mutually Exclusive Mutations''' | ||
| + | !'''Diagnostic Significance (Yes, No or Unknown)''' | ||
| + | !Prognostic Significance (Yes, No or Unknown) | ||
| + | !Therapeutic Significance (Yes, No or Unknown) | ||
| + | !Notes | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations | ||
| − | + | <span class="blue-text">EXAMPLE:</span> | |
| − | |||
| − | |||
| − | |||
| − | + | EGFR; Exon 20 mutations | |
| − | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations | |
| + | |<span class="blue-text">EXAMPLE:</span> TSG | ||
| + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | ||
| − | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) | |
| + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H | ||
| + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). | ||
| + | <br /> | ||
| + | |} | ||
| + | Note: A more extensive list of mutations can be found in cBioportal (https://www.cbioportal.org/), COSMIC (https://cancer.sanger.ac.uk/cosmic), ICGC (https://dcc.icgc.org/) and/or other databases. When applicable, gene-specific pages within the CCGA site directly link to pertinent external content. | ||
| − | |||
| − | |||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Gene Mutations (SNV/INDEL)|The content below was from the old template. Please incorporate above.}} | |
| − | + | A few genes were noted to be altered during the transformed stages of CML, namely TP53'', RB1, MYC, CDKN2A, NRAS, KRAS, RUNX1, MECOM, TET2, CBL, ASXL1, IDH1'' and ''IDH2''. | |
| − | |||
| − | |||
| − | |||
| − | == | + | </blockquote> |
| − | + | ==Epigenomic Alterations== | |
| − | + | Not Applicable. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Genes and Main Pathways Involved== | |
| − | == | + | Put your text here and fill in the table <span style="color:#0070C0">(''Instructions: Can include references in the table. Do not delete table.'')</span> |
| − | + | {| class="wikitable sortable" | |
| + | |- | ||
| + | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''BRAF'' and ''MAP2K1''; Activating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling | ||
| + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''CDKN2A''; Inactivating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation | ||
| + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> ''KMT2C'' and ''ARID1A''; Inactivating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling | ||
| + | |<span class="blue-text">EXAMPLE:</span> | ||
| + | {| class="wikitable sortable" | ||
| + | |- | ||
| + | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling | ||
| + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation | ||
| + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division | ||
| + | |- | ||
| + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations | ||
| + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling | ||
| + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program | ||
| + | |} | ||
| − | + | <blockquote class='blockedit'>{{Box-round|title=v4:Genes and Main Pathways Involved|The content below was from the old template. Please incorporate above.}} | |
| − | + | [[File:BCR-ABL-ASS1 abnormal double fusions.jpg|thumb|Image courtesy of Fabiola Quintero-Rivera, MD]] | |
| − | + | Breakpoint Cluster Region protein (BCR) is encoded by ''BCR'' gene, located on chromosome 22. BCR protein has serine/threonine kinase activity.<ref name=":0" /> The protein is also a GTPase-activating protein for p21rac and other kinases.<ref>{{Cite journal|title=BCR BCR activator of RhoGEF and GTPase [Homo sapiens (human)] - Gene - NCBI|url=https://www.ncbi.nlm.nih.gov/gene?cmd=Retrieve&dopt=full_report&list_uids=613}}</ref> BCR protein is involved in the two main pathways: FGFR1 mutant receptor activation and G-protein signaling HRAS regulation pathway.<ref>{{Cite journal|last=Mn|first=Peiris|last2=F|first2=Li|last3=Dj|first3=Donoghue|date=2019|title=BCR: A Promiscuous Fusion Partner in Hematopoietic Disorders|url=https://pubmed.ncbi.nlm.nih.gov/31105873/|language=en|doi=10.18632/oncotarget.26837|pmc=PMC6505627|pmid=31105873}}</ref> BCR-associated genetic rearrangement gives rise to hematological disorders. The ''ABL1'' gene is located on chromosome 9q34.12 and encodes for ABL1 protein, which was discovered to be a tyrosine kinase protein.<ref>{{Cite journal|last=B|first=Chereda|last2=Jv|first2=Melo|date=2015|title=Natural Course and Biology of CML|url=https://pubmed.ncbi.nlm.nih.gov/25814077/|language=en|pmid=25814077}}</ref> Depending on the breakpoint of the ''BCR'' gene, the size of the fusion protein can vary: p190bcr-abl, p210bcr-abl, and p230bcr-abl, leading to three different isoforms.<ref name=":0" /> ''BCR-ABL1'' gene fusion encodes a chimeric protein, which is mostly 210 kDa(P210''BCRABL1'') with constitutive tyrosine-kinase activity, escaping the cytokine regulation and regulatory controls of many intracellular signaling pathways that are associated with proliferation, differentiation and apoptosis.<ref>{{Cite journal|last=Jb|first=Konopka|last2=Sm|first2=Watanabe|last3=On|first3=Witte|date=1984|title=An Alteration of the Human C-Abl Protein in K562 Leukemia Cells Unmasks Associated Tyrosine Kinase Activity|url=https://pubmed.ncbi.nlm.nih.gov/6204766/|language=en|pmid=6204766}}</ref><ref>{{Cite journal|last=R|first=Ren|date=2005|title=Mechanisms of BCR-ABL in the Pathogenesis of Chronic Myelogenous Leukaemia|url=https://pubmed.ncbi.nlm.nih.gov/15719031/|language=en|pmid=15719031}}</ref> Many of the target proteins that are affected by dimerization of constitutive kinase activity of BCR-ABL fusion protein include STAT, RAS, RAF, JUN kinase, MYC, AKT, and other transducers.<ref>{{Cite journal|last=S|first=Faderl|last2=M|first2=Talpaz|last3=Z|first3=Estrov|last4=S|first4=O'Brien|last5=R|first5=Kurzrock|last6=Hm|first6=Kantarjian|date=1999|title=The Biology of Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/10403855/|language=en|pmid=10403855}}</ref><ref>{{Cite journal|last=Cl|first=Sawyers|date=1999|title=Chronic Myeloid Leukemia|url=https://pubmed.ncbi.nlm.nih.gov/10219069/|language=en|pmid=10219069}}</ref> It was shown that when CML progresses to the blastic crisis phase, a new additional mutation is acquired GSK3beta, which leads to the activation of beta-catenin, preventing myeloid cell lineages to mature.<ref>{{Cite journal|last=Ch|first=Jamieson|last2=Le|first2=Ailles|last3=Sj|first3=Dylla|last4=M|first4=Muijtjens|last5=C|first5=Jones|last6=Jl|first6=Zehnder|last7=J|first7=Gotlib|last8=K|first8=Li|last9=Mg|first9=Manz|date=2004|title=Granulocyte-macrophage Progenitors as Candidate Leukemic Stem Cells in Blast-Crisis CML|url=https://pubmed.ncbi.nlm.nih.gov/15306667/|language=en|pmid=15306667}}</ref><ref>{{Cite journal|last=Ae|first=Abrahamsson|last2=I|first2=Geron|last3=J|first3=Gotlib|last4=Kh|first4=Dao|last5=Cf|first5=Barroga|last6=Ig|first6=Newton|last7=Fj|first7=Giles|last8=J|first8=Durocher|last9=Rs|first9=Creusot|date=2009|title=Glycogen Synthase Kinase 3beta Missplicing Contributes to Leukemia Stem Cell Generation|url=https://pubmed.ncbi.nlm.nih.gov/19237556/|language=en|doi=10.1073/pnas.0900189106|pmc=PMC2646624|pmid=19237556}}</ref> | |
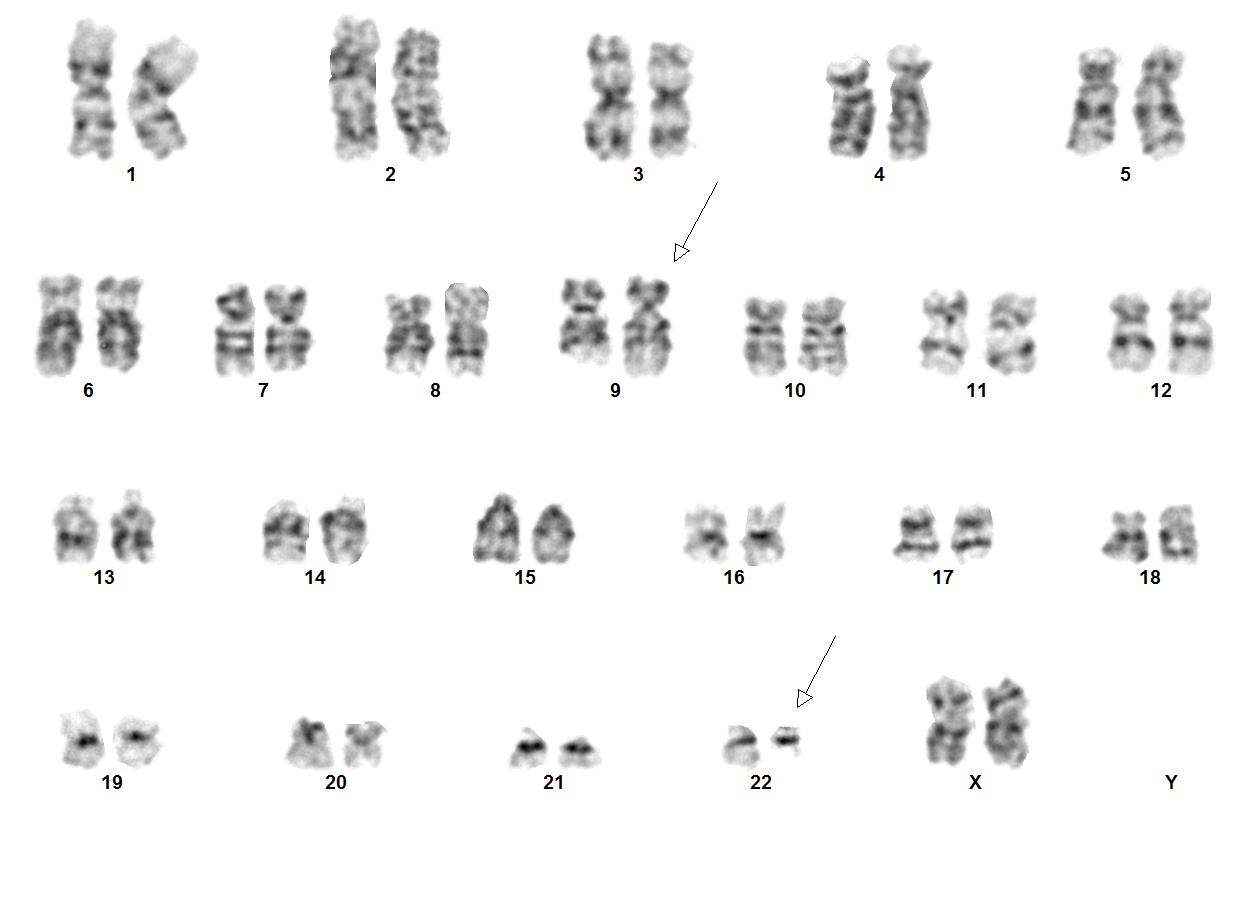
| + | [[File:9;22 image2K Abnormal Karyogram.jpg|thumb|Image courtesy of Fabiola Quintero-Rivera, MD]] | ||
| − | + | </blockquote> | |
| + | ==Genetic Diagnostic Testing Methods== | ||
| + | Majority of the patients with CML are initially diagnosed through a blood test - complete blood count - before clinical manifestations. Bone marrow aspirate and core biopsy are performed sequentially to further support the process of making diagnosis as well as assessing percentage of blasts and basophils.<ref>{{Cite journal|last=E|first=Jabbour|last2=H|first2=Kantarjian|date=2018|title=Chronic Myeloid Leukemia: 2018 Update on Diagnosis, Therapy and Monitoring|url=https://pubmed.ncbi.nlm.nih.gov/29411417/|language=en|pmid=29411417}}</ref> Histologic features of CML in the peripheral blood are helpful in deriving the CML diagnosis. Ancillary tests are performed to clinch the diagnosis: immunohistochemistry, flow cytometry, conventional cytogenetic analysis, FISH and molecular RT-PCR-based studies. | ||
| − | + | Cytogenetic testing is used in CML patients to monitor how patients are responding to the treatment by detecting the number of cells with the Philadelphia chromosome. In conjunction with cytogenetic testing, FISH and PCR are ordered to quantify treatment monitoring processes in CML. FISH allows the detection of ''BCR-ABL'' gene, which is essentially considered to be more of a sensitive test compared to cytogenetic testing. PCR is performed to find ''BCR-ABL'' fusion gene and other molecular abnormalities. PCR is very efficient because it can detect even one abnormal cell from approximately 1 million healthy cells. FISH can be used in rare cases where molecular transcripts are not detected. | |
| − | + | Continuous monitoring is part of the standard of care in CML patients because it allows the clinicians to identify treatment failure, disease evolution and drug regimen adherence. Minimal residual disease monitoring is done by using RT-PCR. Major molecular response (MMR) is the critical goal of CML treatment. Complete or deep molecular response is achieved when there is absence of BCR-ABL1 transcript or >4.5 logs below baseline level.<ref>{{Cite journal|last=gknation|date=2015|title=Treatment Outcomes|url=https://www.lls.org/leukemia/chronic-myeloid-leukemia/treatment/treatment-outcomes|language=en}}</ref> | |
| − | |||
| − | + | ==Familial Forms== | |
| − | + | Not Applicable. | |
| − | + | ==Additional Information== | |
| − | < | + | Put your text here |
| + | |||
| + | ==Links== | ||
| + | |||
| + | [[ABL1]] | ||
| + | |||
| + | [[BCR]] | ||
| + | |||
| + | ==References== | ||
| + | (use the "Cite" icon at the top of the page) <span style="color:#0070C0">(''Instructions: Add each reference into the text above by clicking on where you want to insert the reference, selecting the “Cite” icon at the top of the page, and using the “Automatic” tab option to search such as by PMID to select the reference to insert. The reference list in this section will be automatically generated and sorted.''</span> <span style="color:#0070C0">''If a PMID is not available, such as for a book, please use the “Cite” icon, select “Manual” and then “Basic Form”, and include the entire reference''</span><span style="color:#0070C0">''.''</span><span style="color:#0070C0">) </span> <references /> | ||
| + | |||
| + | ''' | ||
| + | |||
| + | ==Notes== | ||
| + | <nowiki>*</nowiki>Primary authors will typically be those that initially create and complete the content of a page. If a subsequent user modifies the content and feels the effort put forth is of high enough significance to warrant listing in the authorship section, please contact the CCGA coordinators (contact information provided on the homepage). Additional global feedback or concerns are also welcome. | ||
| + | <nowiki>*</nowiki>''Citation of this Page'': “Chronic myeloid leukaemia”. Compendium of Cancer Genome Aberrations (CCGA), Cancer Genomics Consortium (CGC), updated {{REVISIONMONTH}}/{{REVISIONDAY}}/{{REVISIONYEAR}}, <nowiki>https://ccga.io/index.php/HAEM5:Chronic_myeloid_leukaemia</nowiki>. | ||
| + | [[Category:HAEM5]][[Category:DISEASE]][[Category:Diseases C]] | ||
Latest revision as of 13:04, 1 May 2024
Haematolymphoid Tumours (5th ed.)
| This page is under construction |
editHAEM5 Conversion NotesThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Chronic Myeloid Leukemia (CML), BCR-ABL1 Positive.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click nearby within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Jack Reid, MD (University of California, Irvine)
Mark Evans, MD (University of California, Irvine)
Fabiola Quintero-Rivera, MD (University of California, Irvine)
Cancer Category / Type
Myeloproliferative neoplasm
Cancer Sub-Classification / Subtype
Not applicable.
Definition / Description of Disease
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm that is characterized by clonal expansion of predominantly granulocytic proliferation (neutrophils, eosinophils and basophils). The majority of the patients with CML are known to have a gene rearrangement called the Philadelphia Chromosome, which is a balanced genetic translocation t(9;22)(q34.1;q11.2) involving a fusion of the Abelson gene (ABL1) from chromosome 9q34 with the breakpoint cluster region (BCR) gene on chromosome 22q11.2. Althought 80% of the clonal evolution in CML cases can be attributed to classic Ph chromosome, secondary cytogenetic aberrations can be seen such as isochromosome 17q, gain of chromosome 8 or 19. ML was first recognized in 1845[1]. Nowell and Hungerford in 1960, who coined the term Philadelphia Chromosome after realizing consistent chromosomal abnormality in leukemic cells.[2] Later in 1973, the characteristic cytogenetic feature of CML was identified: reciprocal translocation of t(9;22)(q34.1;q11.2).[3] CML has the capacity to expand in both myeloid and lymphoid lineages. However, expansion is predominantly in the granulocyte compartment of the myeloid lineages in the bone marrow.[4] In the previous WHO 4th edition, CML was be divided into 3 phases of disease: chronic phase, accelerated phase and blastic phase. Currently, in the revised classification of CML, AP at diagnosis or during treatment has been omitted and replaced by recognising only the chronic and blast phases
Synonyms / Terminology
Formerly chronic myelogenous leukemia or chronic granulocytic leukemia
Epidemiology / Prevalence
Global annual incidence of CML is 1-2 cases per 100,000 population. CML has male predominance with male to female ratio of 1.4:1. The prevalence of CML is increasing due to successful Tyrosine kinase inhibitor (TKI) therapy. Predilection of CML among certain ethnic groups has not been reported.[5] Regional variations in age at diagnosis and overall survival among patients with chronic myeloid leukemia from low and middle income countries.[6] The median age of patients diagnosed with CML is 66 years according to Surveillance, Epidemiology, and End Results (SEER) program and Medical Research Council (MRC) data. Although the etiology of CML is largely unknown, cases of CML have been reported in association with radiation exposure. No studies have shown any genetic inheritance of CML.
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Lymphadenopathy (uncommon) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE:
Sites of InvolvementSpleen is known to be the most common site of involvement as patients with CML usually present with splenomegaly. Literature has shown that bone marrow is always involved in patients with CML. Morphologic FeaturesMorphologically, peripheral blood smear shows the classic features of chronic-phase CML: granulocytic leukocytosis with left shift, neutrophilia, no increase in blasts, myelocyte “bulge” and basophilia.[9] Granulocytes seen in CML patients usually lack dysplastic features. In bone marrow, the aspirate smear shows classic features of CML: marked increase in neutrophils and precursors with a myeloid: erythroid ratio of > 10:1 and small hypolobated megakaryocytes. CML can sometimes present with thrombocytosis in peripheral blood smear, mimicking essential thrombocythemia (ET). Most of the time, primary myelofibrosis (PMF) share overlapping morphological features with CML in the peripheral blood; one distinguishing feature is that the megakaryocyte morphology in PMF is large, bizarre, and hyperchromatic, which is the feature not seen in the CML. ImmunophenotypeThe role of immunohistochemistry is minimal in diagnosing CML. CML usually shows blastic markers (CD 34, CD 117, TdT), which can be useful to confirm extramedullary (splenic involvement with blastic transformation). Lineage-specific markers (MPO, lysosome, CD42b, CD79a, PAX5, CD3) are helpful in distinguishing among myeloid, lymphoid or megakaryocytic transformation.
Chromosomal Rearrangements (Gene Fusions)Put your text here and fill in the table
Individual Region Genomic Gain / Loss / LOHPut your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
|