Difference between revisions of "Test"
| [checked revision] | [checked revision] |
Bailey.Glen (talk | contribs) |
Bailey.Glen (talk | contribs) |
||
| Line 54: | Line 54: | ||
{| class="wikitable" | {| class="wikitable" | ||
|'''Signs and Symptoms''' | |'''Signs and Symptoms''' | ||
| − | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) | + | |<span class="blue-text">EXAMPLE:</span> Asymptomatic (incidental finding on complete blood counts) |
| − | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) | + | <span class="blue-text">EXAMPLE:</span> B-symptoms (weight loss, fever, night sweats) |
| − | <span class="blue-text">EXAMPLE:</span> Fatigue | + | <span class="blue-text">EXAMPLE:</span> Fatigue |
| − | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) | + | <span class="blue-text">EXAMPLE:</span> Lymphadenopathy (uncommon) |
|- | |- | ||
|'''Laboratory Findings''' | |'''Laboratory Findings''' | ||
| − | |<span class="blue-text">EXAMPLE:</span> Cytopenias | + | |<span class="blue-text">EXAMPLE:</span> Cytopenias |
| − | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) | + | <span class="blue-text">EXAMPLE:</span> Lymphocytosis (low level) |
|} | |} | ||
| Line 110: | Line 110: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> t(9;22)(q34;q11.2)||<span class="blue-text">EXAMPLE:</span> 3'ABL1 / 5'BCR||<span class="blue-text">EXAMPLE:</span> der(22)||<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | <span class="blue-text">EXAMPLE:</span> 30% (add reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add reference) |
|Yes | |Yes | ||
|No | |No | ||
|Yes | |Yes | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | The t(9;22) is diagnostic of CML in the appropriate morphology and clinical context (add reference). This fusion is responsive to targeted therapy such as Imatinib (Gleevec) (add reference). | ||
| Line 196: | Line 196: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
7 | 7 | ||
| − | |<span class="blue-text">EXAMPLE:</span> Loss | + | |<span class="blue-text">EXAMPLE:</span> Loss |
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
chr7:1- 159,335,973 [hg38] | chr7:1- 159,335,973 [hg38] | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
chr7 | chr7 | ||
| Line 209: | Line 209: | ||
|Yes | |Yes | ||
|No | |No | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | Presence of monosomy 7 (or 7q deletion) is sufficient for a diagnosis of AML with MDS-related changes when there is ≥20% blasts and no prior therapy (add reference). Monosomy 7/7q deletion is associated with a poor prognosis in AML (add reference). | ||
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
8 | 8 | ||
| − | |<span class="blue-text">EXAMPLE:</span> Gain | + | |<span class="blue-text">EXAMPLE:</span> Gain |
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
chr8:1-145,138,636 [hg38] | chr8:1-145,138,636 [hg38] | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
chr8 | chr8 | ||
| Line 226: | Line 226: | ||
|No | |No | ||
|No | |No | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
Common recurrent secondary finding for t(8;21) (add reference). | Common recurrent secondary finding for t(8;21) (add reference). | ||
| Line 261: | Line 261: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
Co-deletion of 1p and 18q | Co-deletion of 1p and 18q | ||
| Line 267: | Line 267: | ||
|No | |No | ||
|No | |No | ||
| − | |<span class="blue-text">EXAMPLE:</span> | + | |<span class="blue-text">EXAMPLE:</span> |
See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | See chromosomal rearrangements table as this pattern is due to an unbalanced derivative translocation associated with oligodendroglioma (add reference). | ||
| Line 312: | Line 312: | ||
!Notes | !Notes | ||
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations | + | |<span class="blue-text">EXAMPLE:</span> TP53; Variable LOF mutations |
| − | <span class="blue-text">EXAMPLE:</span> | + | <span class="blue-text">EXAMPLE:</span> |
EGFR; Exon 20 mutations | EGFR; Exon 20 mutations | ||
| − | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations | + | <span class="blue-text">EXAMPLE:</span> BRAF; Activating mutations |
| − | |<span class="blue-text">EXAMPLE:</span> TSG | + | |<span class="blue-text">EXAMPLE:</span> TSG |
| − | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) | + | |<span class="blue-text">EXAMPLE:</span> 20% (COSMIC) |
| − | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) | + | <span class="blue-text">EXAMPLE:</span> 30% (add Reference) |
| − | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H | + | |<span class="blue-text">EXAMPLE:</span> IDH1 R123H |
| − | |<span class="blue-text">EXAMPLE:</span> EGFR amplification | + | |<span class="blue-text">EXAMPLE:</span> EGFR amplification |
| | | | ||
| | | | ||
| | | | ||
| − | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). | + | |<span class="blue-text">EXAMPLE:</span> Excludes hairy cell leukemia (HCL) (add reference). |
<br /> | <br /> | ||
|} | |} | ||
| Line 364: | Line 364: | ||
!Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | !Gene; Genetic Alteration!!Pathway!!Pathophysiologic Outcome | ||
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations | + | |<span class="blue-text">EXAMPLE:</span> BRAF and MAP2K1; Activating mutations |
| − | |<span class="blue-text">EXAMPLE:</span> MAPK signaling | + | |<span class="blue-text">EXAMPLE:</span> MAPK signaling |
| − | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation | + | |<span class="blue-text">EXAMPLE:</span> Increased cell growth and proliferation |
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> CDKN2A; Inactivating mutations |
| − | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation | + | |<span class="blue-text">EXAMPLE:</span> Cell cycle regulation |
| − | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division | + | |<span class="blue-text">EXAMPLE:</span> Unregulated cell division |
|- | |- | ||
| − | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations | + | |<span class="blue-text">EXAMPLE:</span> KMT2C and ARID1A; Inactivating mutations |
| − | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling | + | |<span class="blue-text">EXAMPLE:</span> Histone modification, chromatin remodeling |
| − | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program | + | |<span class="blue-text">EXAMPLE:</span> Abnormal gene expression program |
|} | |} | ||
Latest revision as of 13:04, 1 May 2024
Haematolymphoid Tumours (5th ed.)
| This page is under construction |
editHAEM5 Conversion NotesThis page was converted to the new template on 2023-12-07. The original page can be found at HAEM4:Chronic Myeloid Leukemia (CML), BCR-ABL1 Positive.
(General Instructions – The main focus of these pages is the clinically significant genetic alterations in each disease type. Use HUGO-approved gene names and symbols (italicized when appropriate), HGVS-based nomenclature for variants, as well as generic names of drugs and testing platforms or assays if applicable. Please complete tables whenever possible and do not delete them (add N/A if not applicable in the table and delete the examples); to add (or move) a row or column to a table, click nearby within the table and select the > symbol that appears to be given options. Please do not delete or alter the section headings. The use of bullet points alongside short blocks of text rather than only large paragraphs is encouraged. Additional instructions below in italicized blue text should not be included in the final page content. Please also see Author_Instructions and FAQs as well as contact your Associate Editor or Technical Support)
Primary Author(s)*
Jack Reid, MD (University of California, Irvine)
Mark Evans, MD (University of California, Irvine)
Fabiola Quintero-Rivera, MD (University of California, Irvine)
Cancer Category / Type
Myeloproliferative neoplasm
Cancer Sub-Classification / Subtype
Not applicable.
Definition / Description of Disease
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm that is characterized by clonal expansion of predominantly granulocytic proliferation (neutrophils, eosinophils and basophils). The majority of the patients with CML are known to have a gene rearrangement called the Philadelphia Chromosome, which is a balanced genetic translocation t(9;22)(q34.1;q11.2) involving a fusion of the Abelson gene (ABL1) from chromosome 9q34 with the breakpoint cluster region (BCR) gene on chromosome 22q11.2. Althought 80% of the clonal evolution in CML cases can be attributed to classic Ph chromosome, secondary cytogenetic aberrations can be seen such as isochromosome 17q, gain of chromosome 8 or 19. ML was first recognized in 1845[1]. Nowell and Hungerford in 1960, who coined the term Philadelphia Chromosome after realizing consistent chromosomal abnormality in leukemic cells.[2] Later in 1973, the characteristic cytogenetic feature of CML was identified: reciprocal translocation of t(9;22)(q34.1;q11.2).[3] CML has the capacity to expand in both myeloid and lymphoid lineages. However, expansion is predominantly in the granulocyte compartment of the myeloid lineages in the bone marrow.[4] In the previous WHO 4th edition, CML was be divided into 3 phases of disease: chronic phase, accelerated phase and blastic phase. Currently, in the revised classification of CML, AP at diagnosis or during treatment has been omitted and replaced by recognising only the chronic and blast phases
Synonyms / Terminology
Formerly chronic myelogenous leukemia or chronic granulocytic leukemia
Epidemiology / Prevalence
Global annual incidence of CML is 1-2 cases per 100,000 population. CML has male predominance with male to female ratio of 1.4:1. The prevalence of CML is increasing due to successful Tyrosine kinase inhibitor (TKI) therapy. Predilection of CML among certain ethnic groups has not been reported.[5] Regional variations in age at diagnosis and overall survival among patients with chronic myeloid leukemia from low and middle income countries.[6] The median age of patients diagnosed with CML is 66 years according to Surveillance, Epidemiology, and End Results (SEER) program and Medical Research Council (MRC) data. Although the etiology of CML is largely unknown, cases of CML have been reported in association with radiation exposure. No studies have shown any genetic inheritance of CML.
Clinical Features
Put your text here and fill in the table (Instruction: Can include references in the table. Do not delete table.)
| Signs and Symptoms | EXAMPLE: Asymptomatic (incidental finding on complete blood counts)
EXAMPLE: B-symptoms (weight loss, fever, night sweats) EXAMPLE: Lymphadenopathy (uncommon) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Laboratory Findings | EXAMPLE: Cytopenias
EXAMPLE:
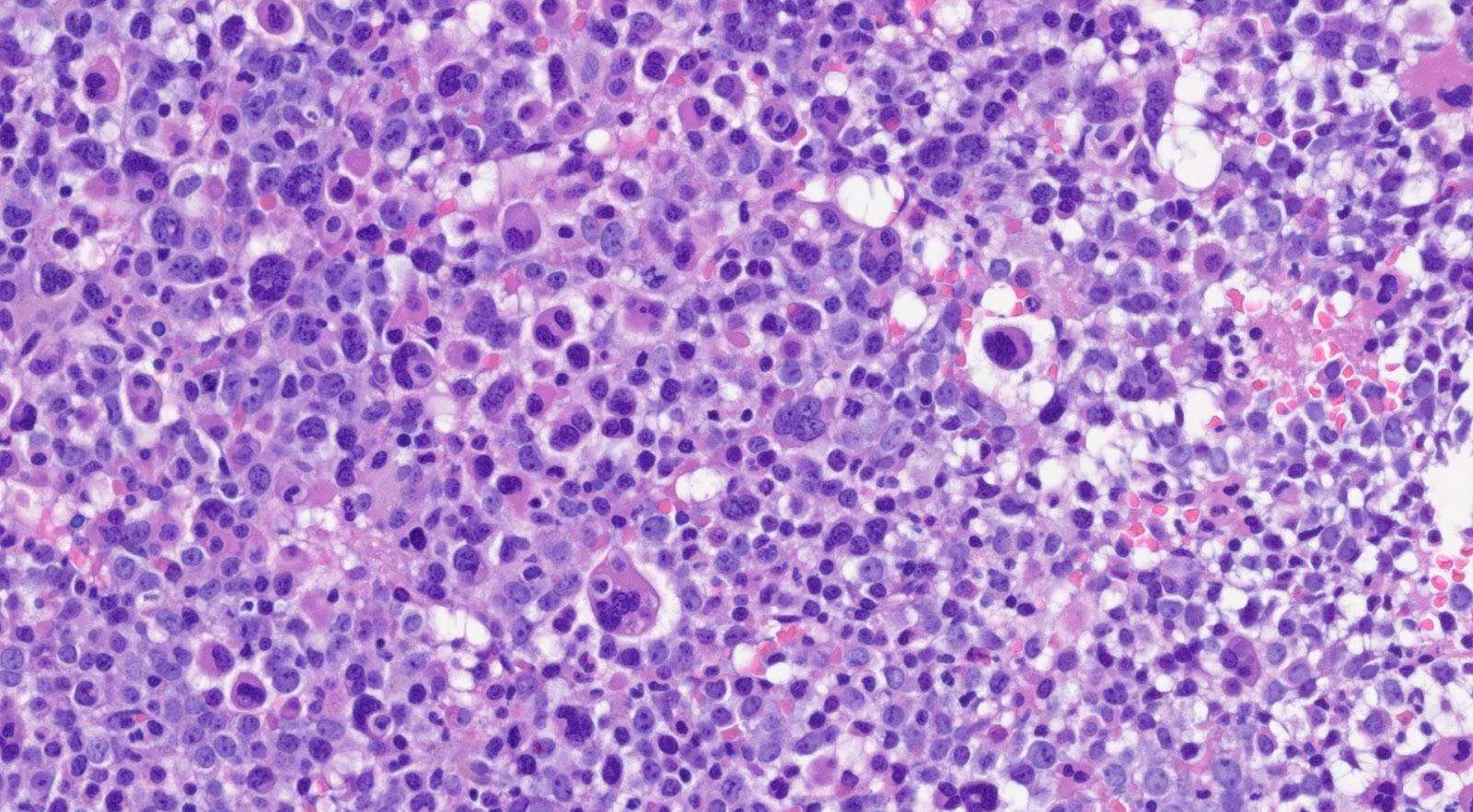
Sites of InvolvementSpleen is known to be the most common site of involvement as patients with CML usually present with splenomegaly. Literature has shown that bone marrow is always involved in patients with CML. Morphologic FeaturesMorphologically, peripheral blood smear shows the classic features of chronic-phase CML: granulocytic leukocytosis with left shift, neutrophilia, no increase in blasts, myelocyte “bulge” and basophilia.[9] Granulocytes seen in CML patients usually lack dysplastic features. In bone marrow, the aspirate smear shows classic features of CML: marked increase in neutrophils and precursors with a myeloid: erythroid ratio of > 10:1 and small hypolobated megakaryocytes. CML can sometimes present with thrombocytosis in peripheral blood smear, mimicking essential thrombocythemia (ET). Most of the time, primary myelofibrosis (PMF) share overlapping morphological features with CML in the peripheral blood; one distinguishing feature is that the megakaryocyte morphology in PMF is large, bizarre, and hyperchromatic, which is the feature not seen in the CML. ImmunophenotypeThe role of immunohistochemistry is minimal in diagnosing CML. CML usually shows blastic markers (CD 34, CD 117, TdT), which can be useful to confirm extramedullary (splenic involvement with blastic transformation). Lineage-specific markers (MPO, lysosome, CD42b, CD79a, PAX5, CD3) are helpful in distinguishing among myeloid, lymphoid or megakaryocytic transformation.
Chromosomal Rearrangements (Gene Fusions)Put your text here and fill in the table
Individual Region Genomic Gain / Loss / LOHPut your text here and fill in the table (Instructions: Includes aberrations not involving gene fusions. Can include references in the table. Can refer to CGC workgroup tables as linked on the homepage if applicable. Do not delete table.)
|